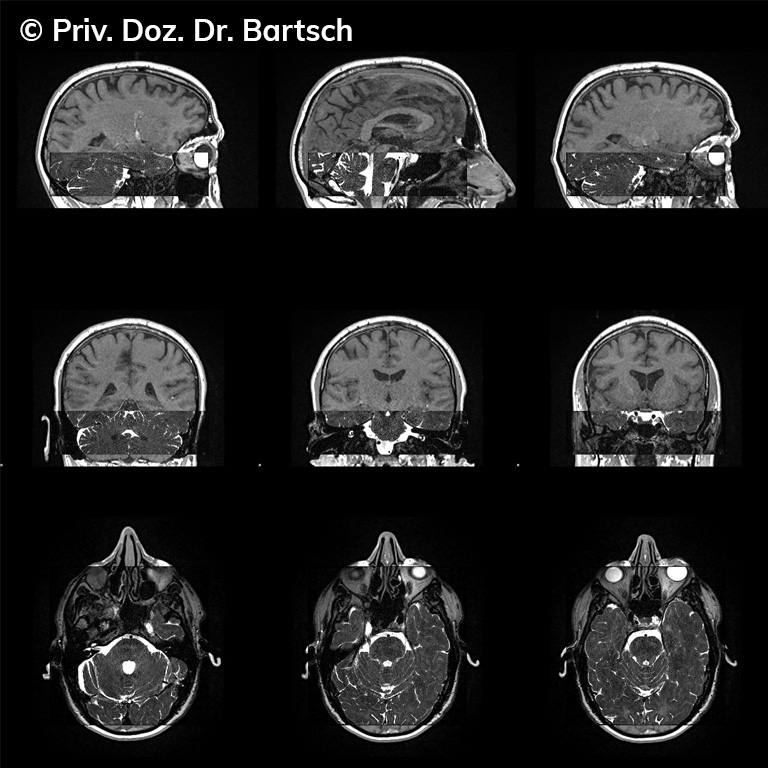
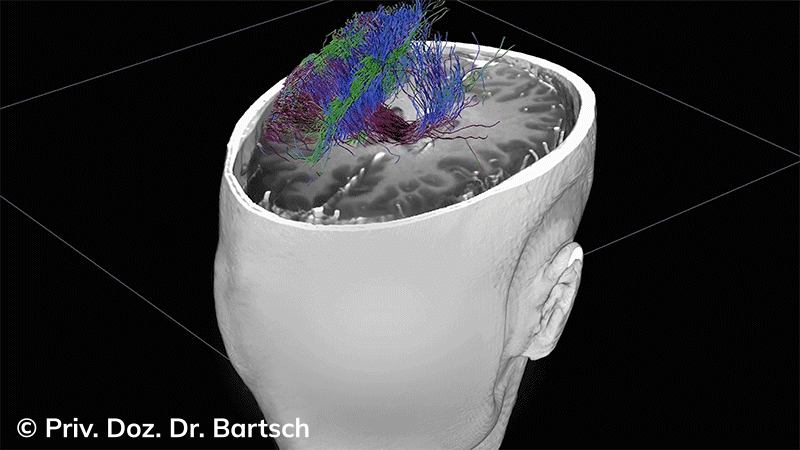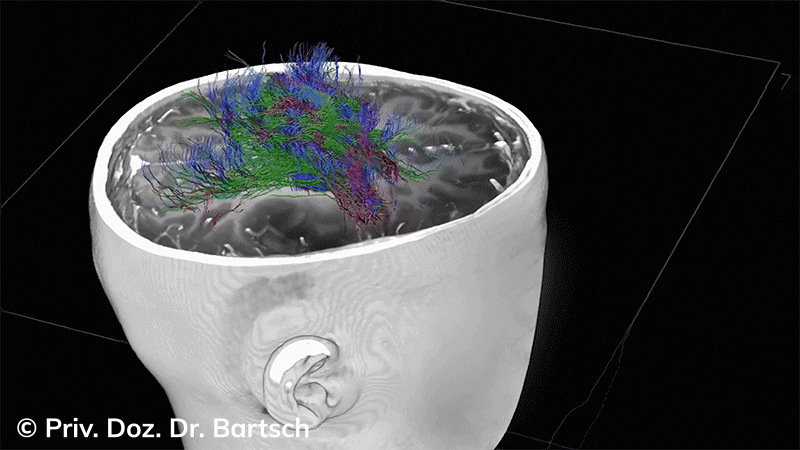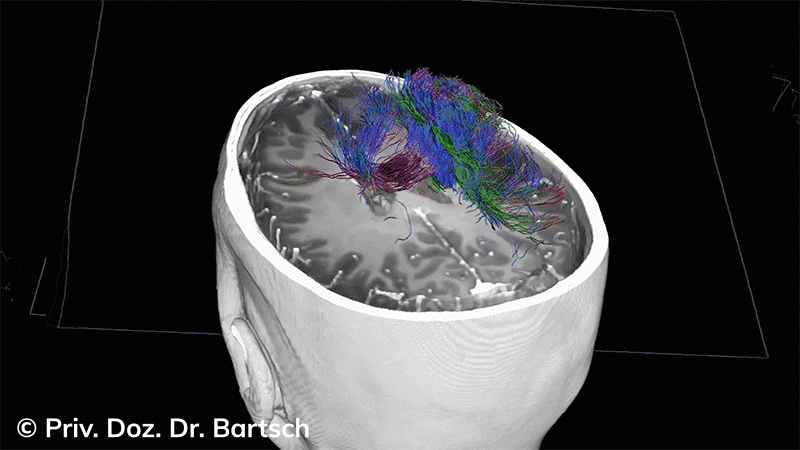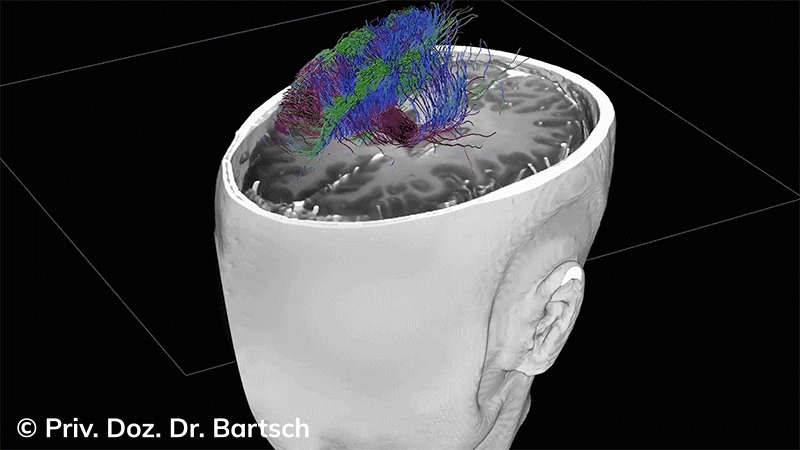
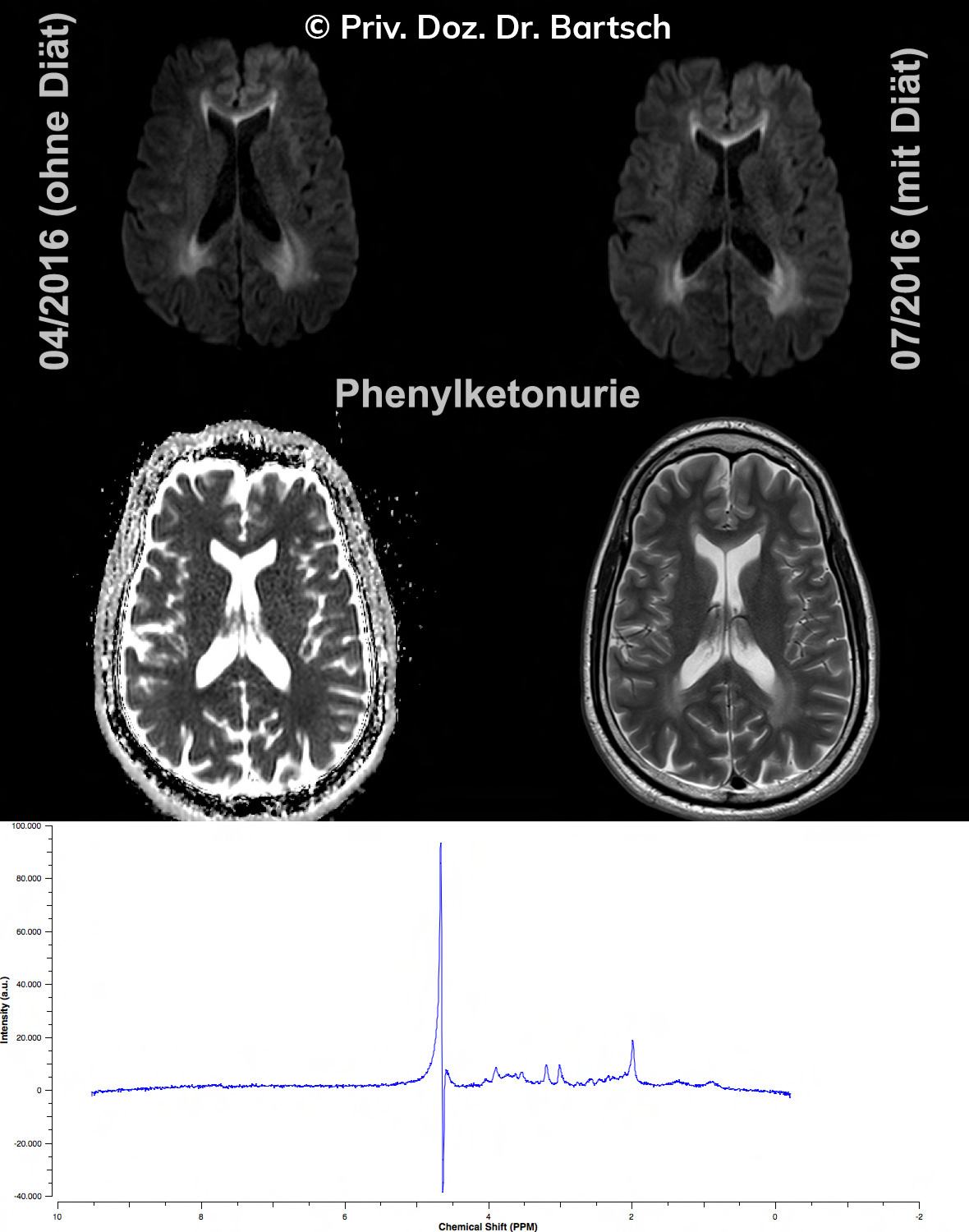
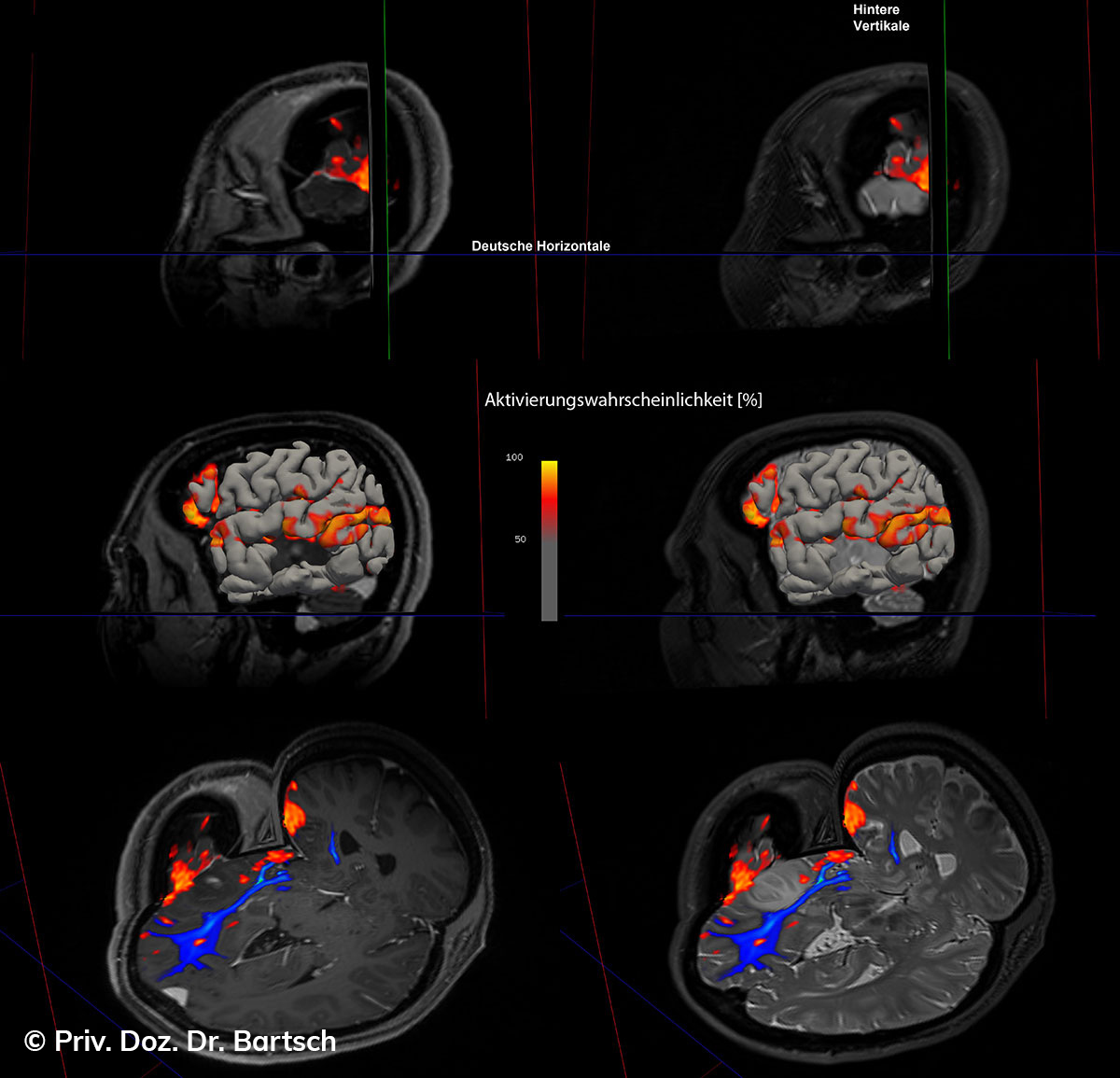
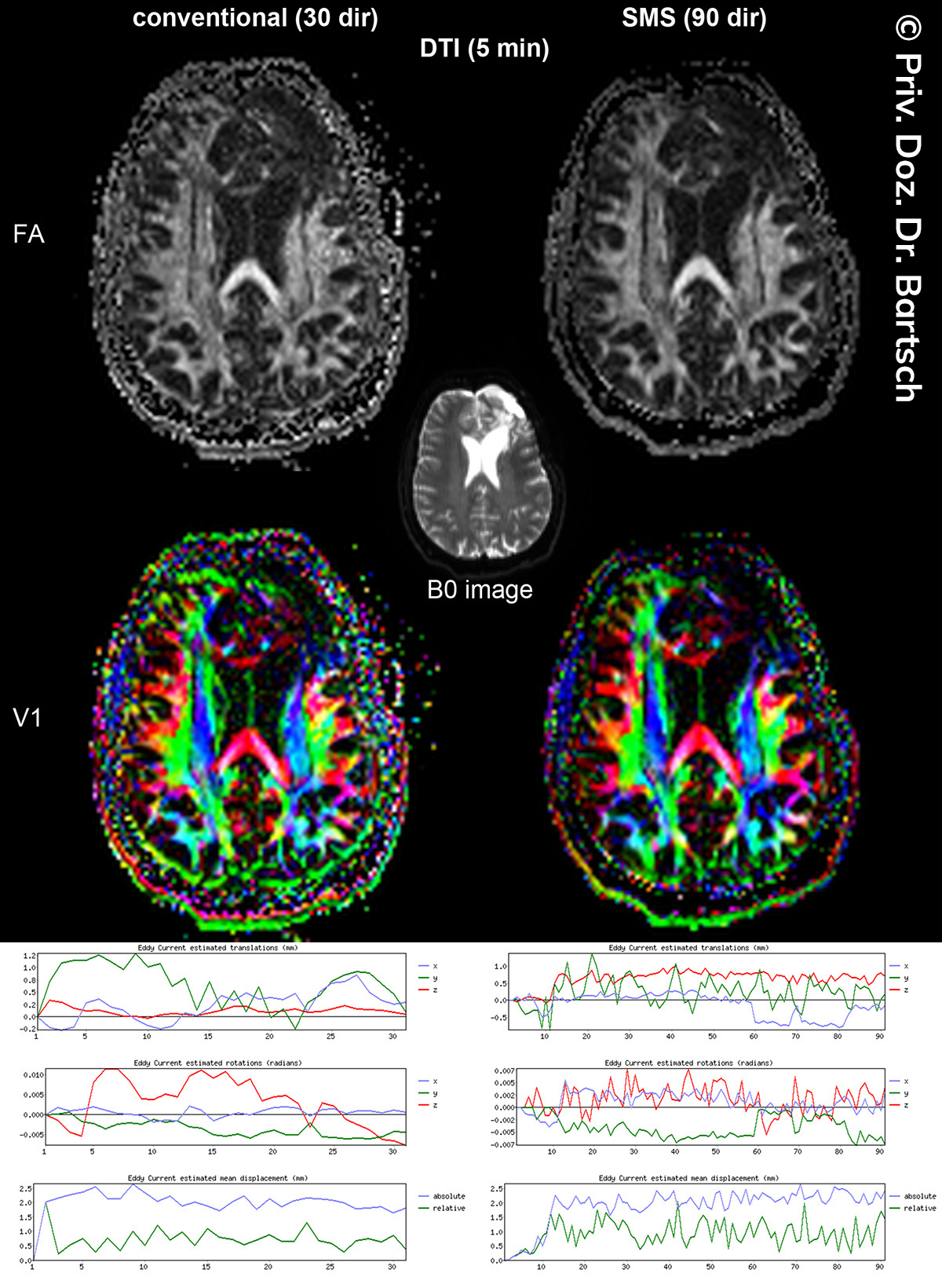
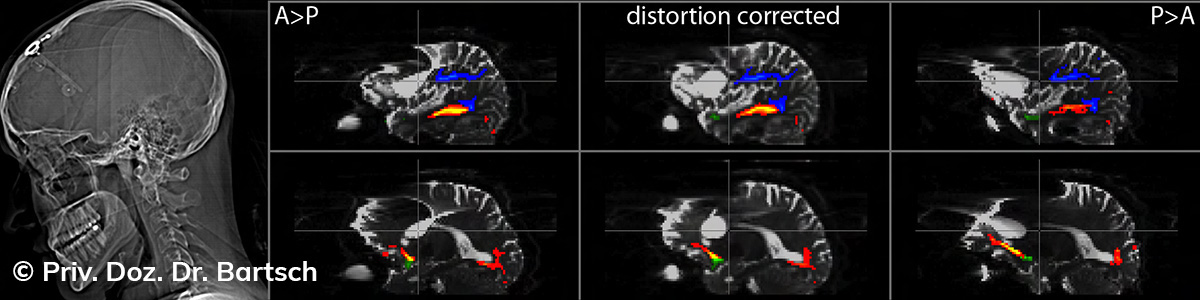
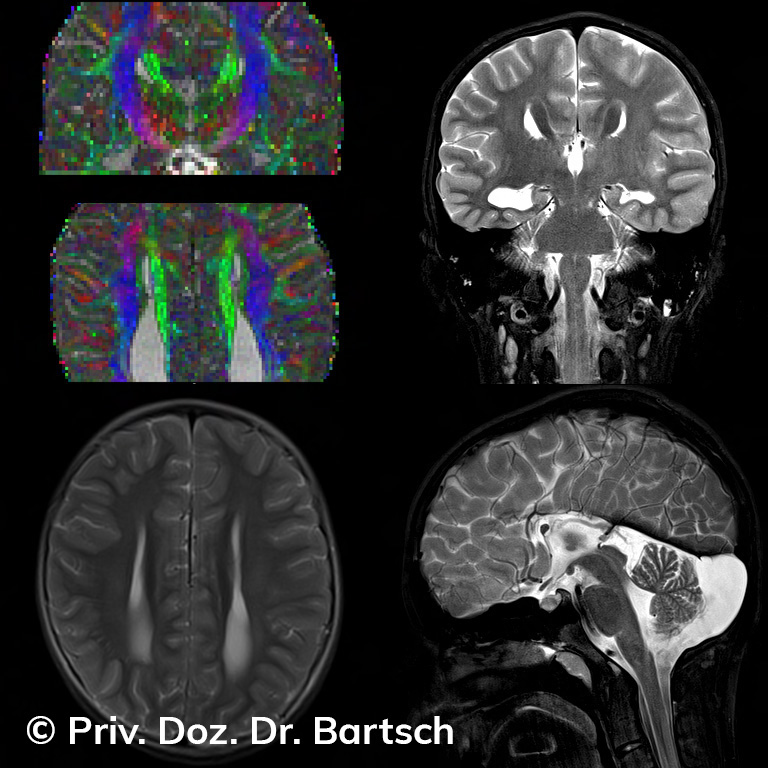
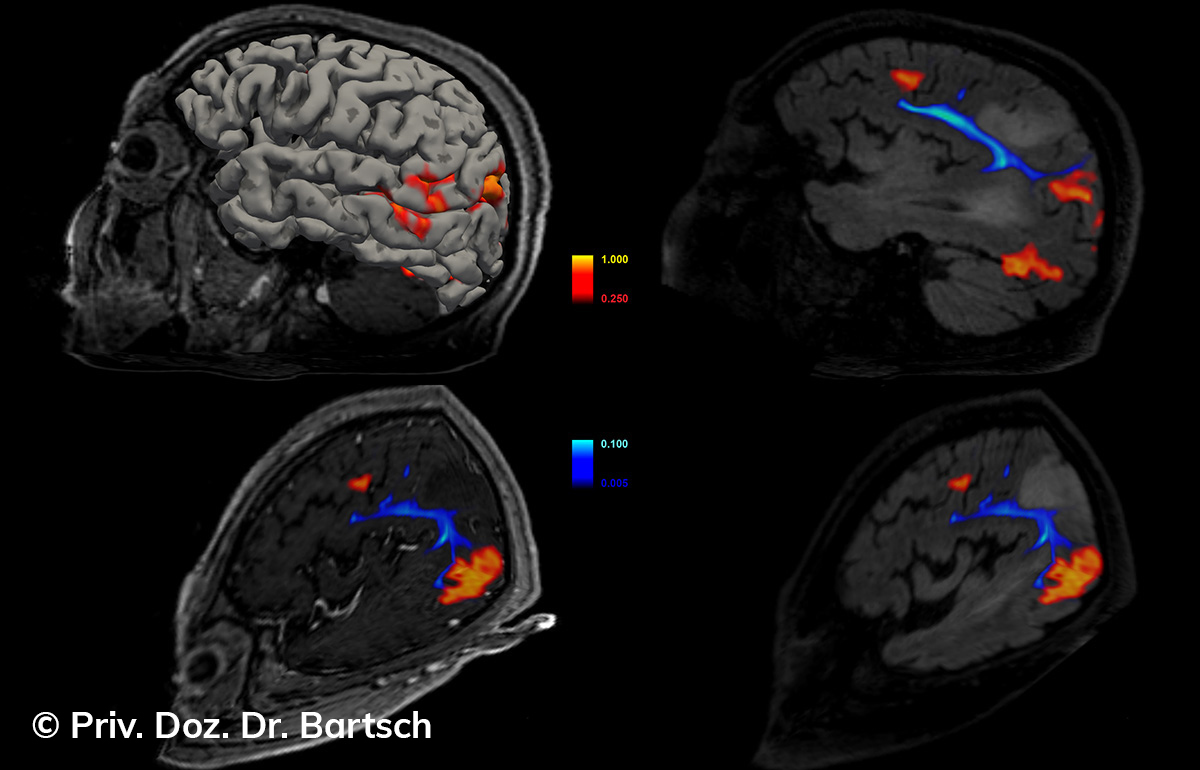
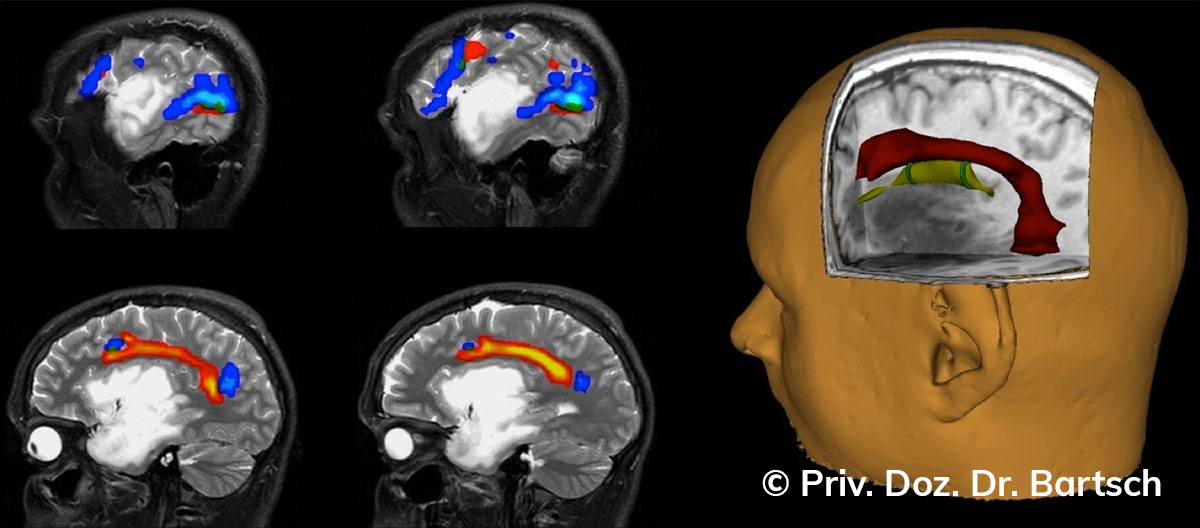
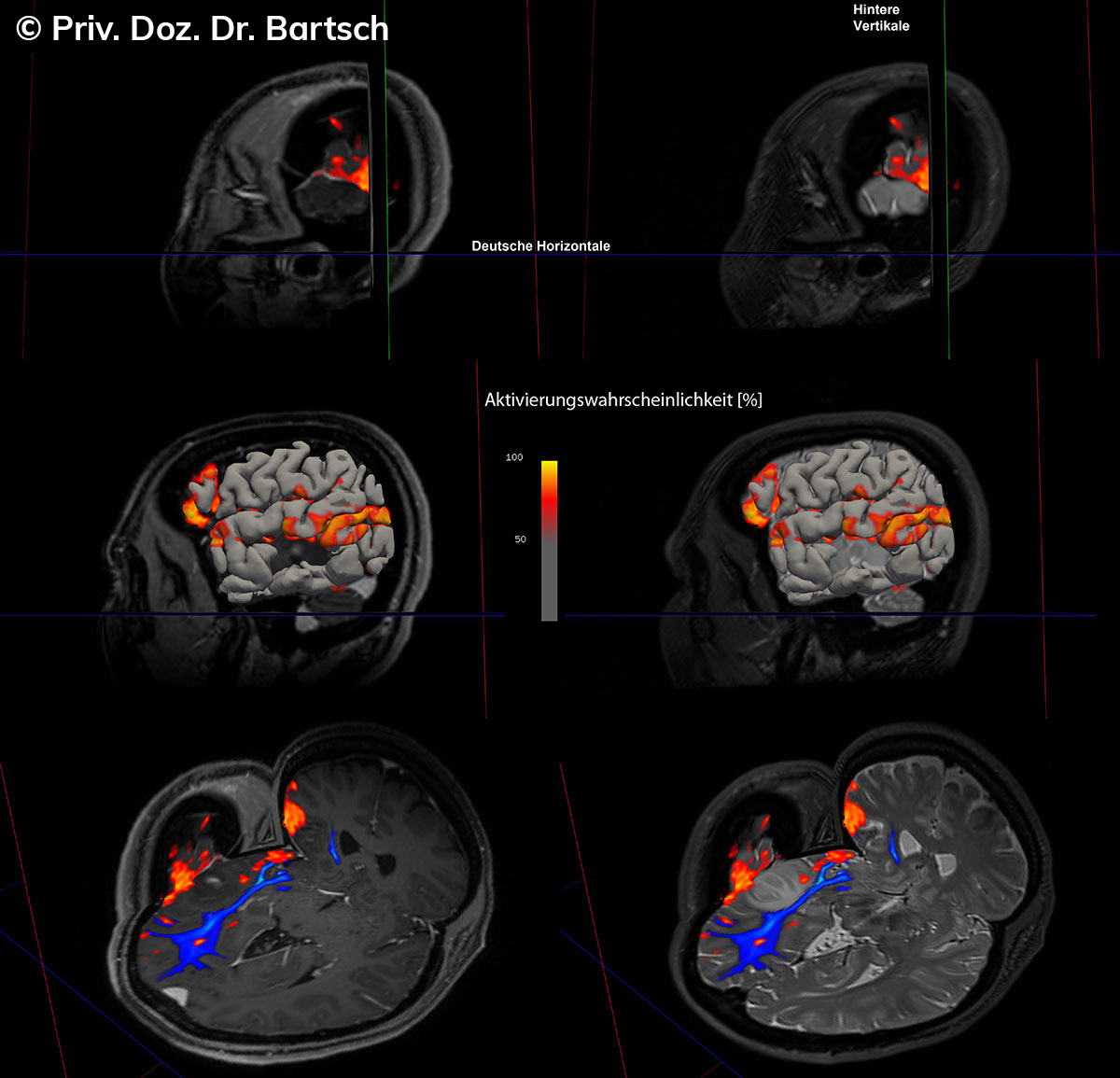
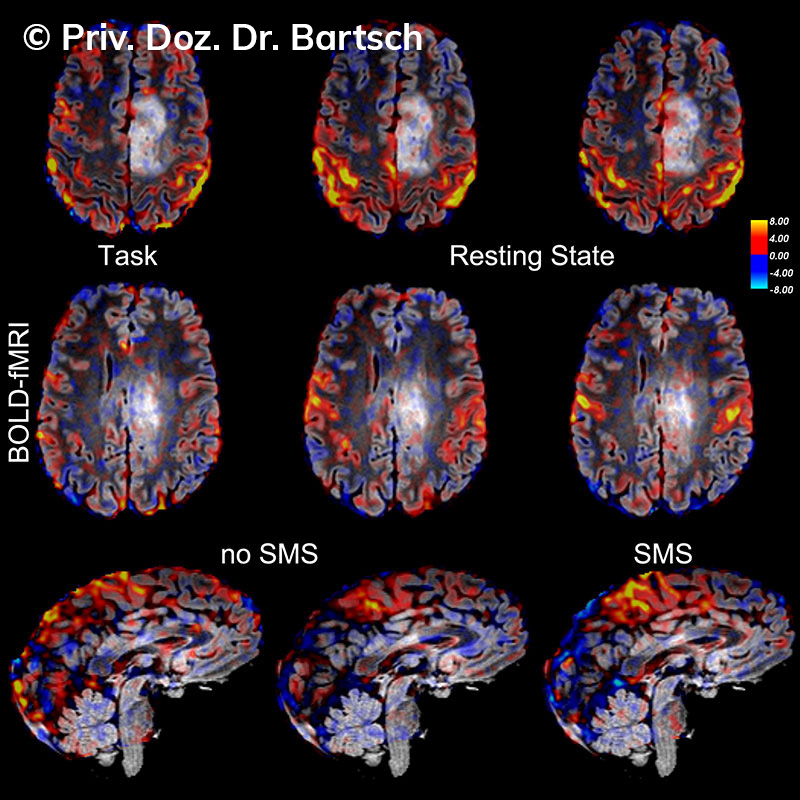
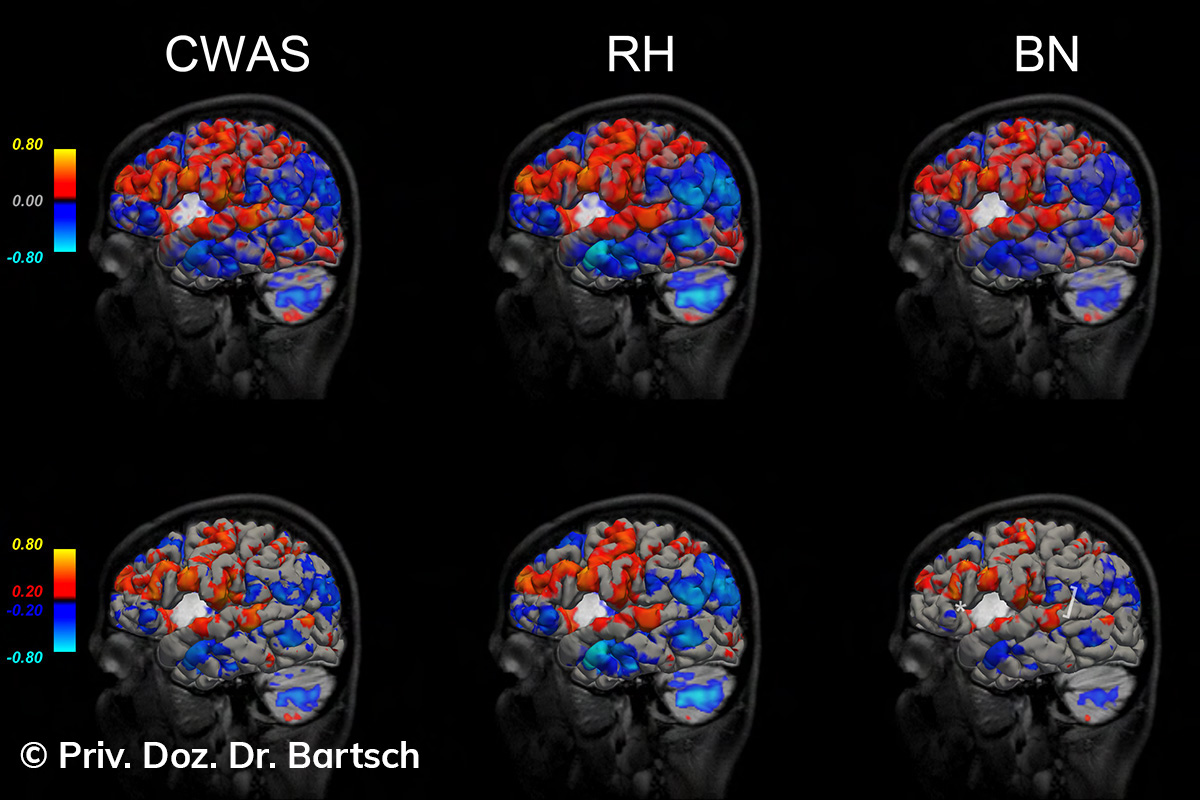
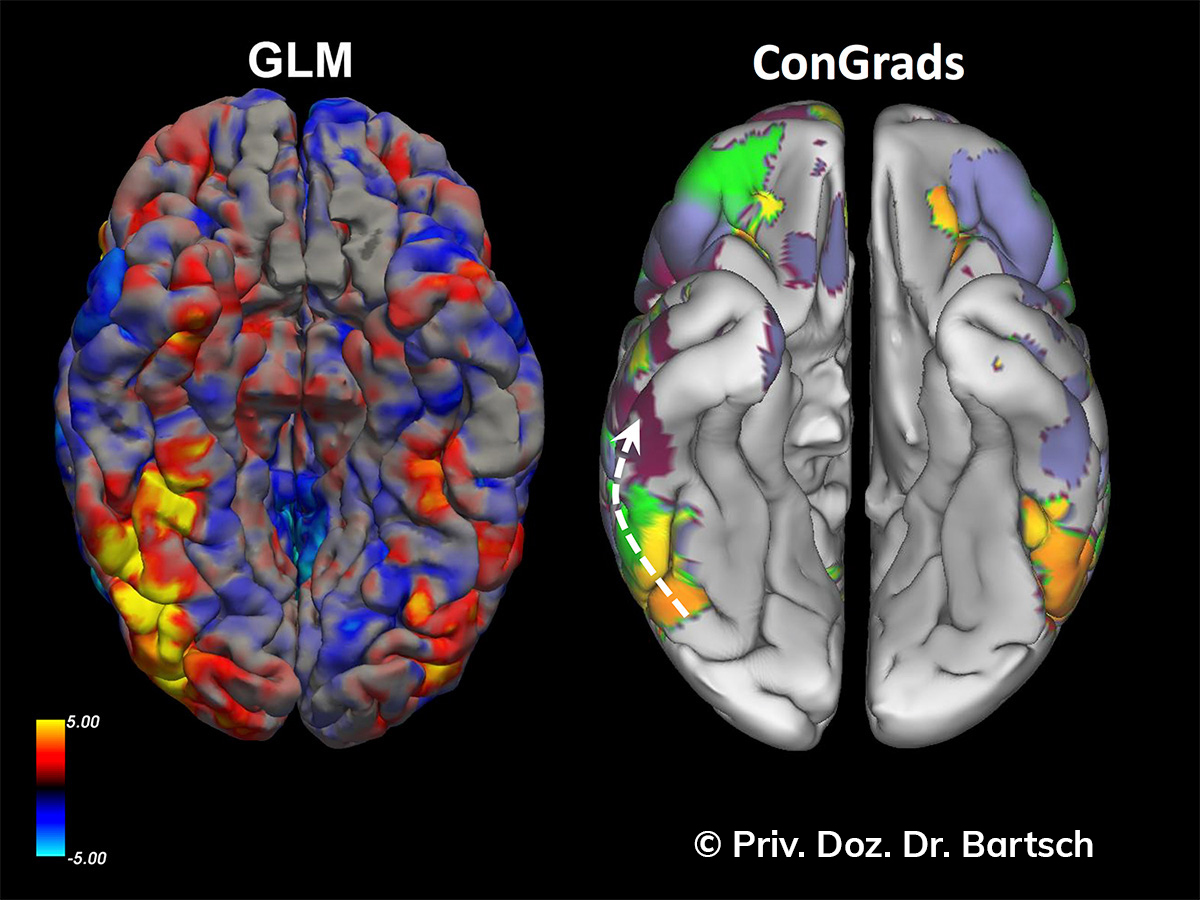
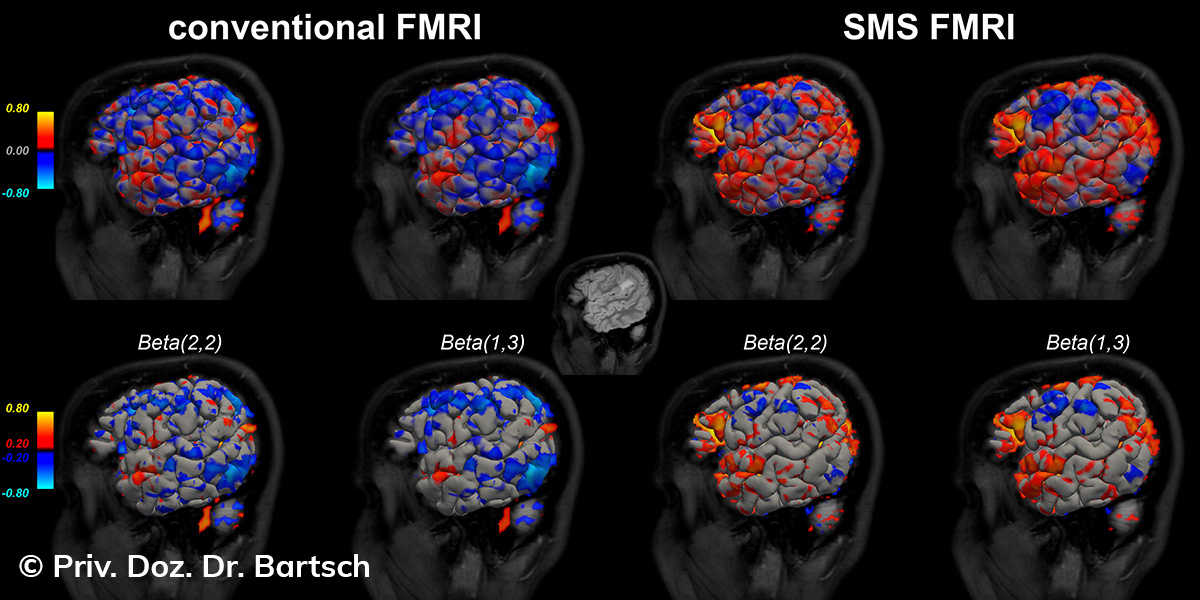
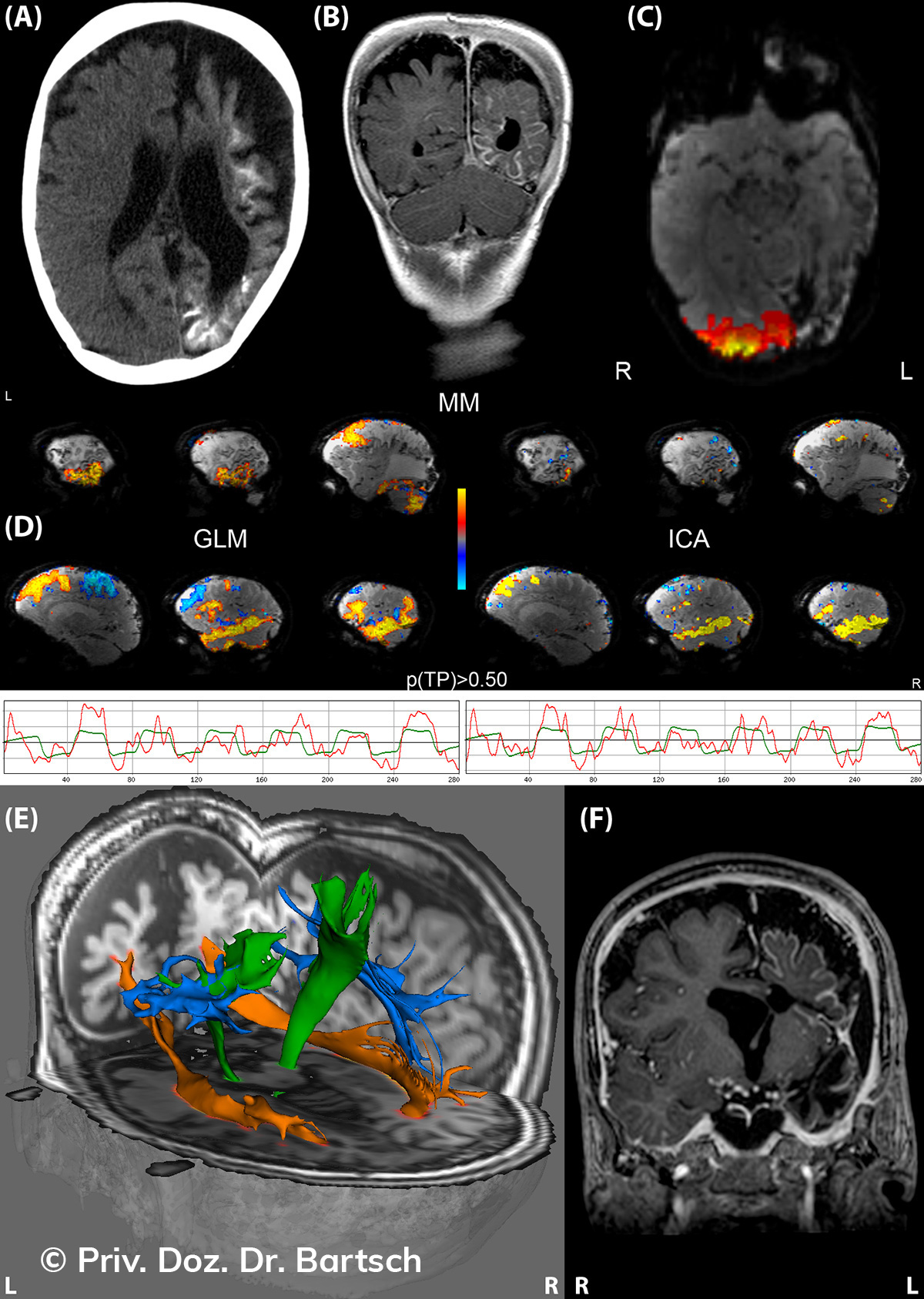
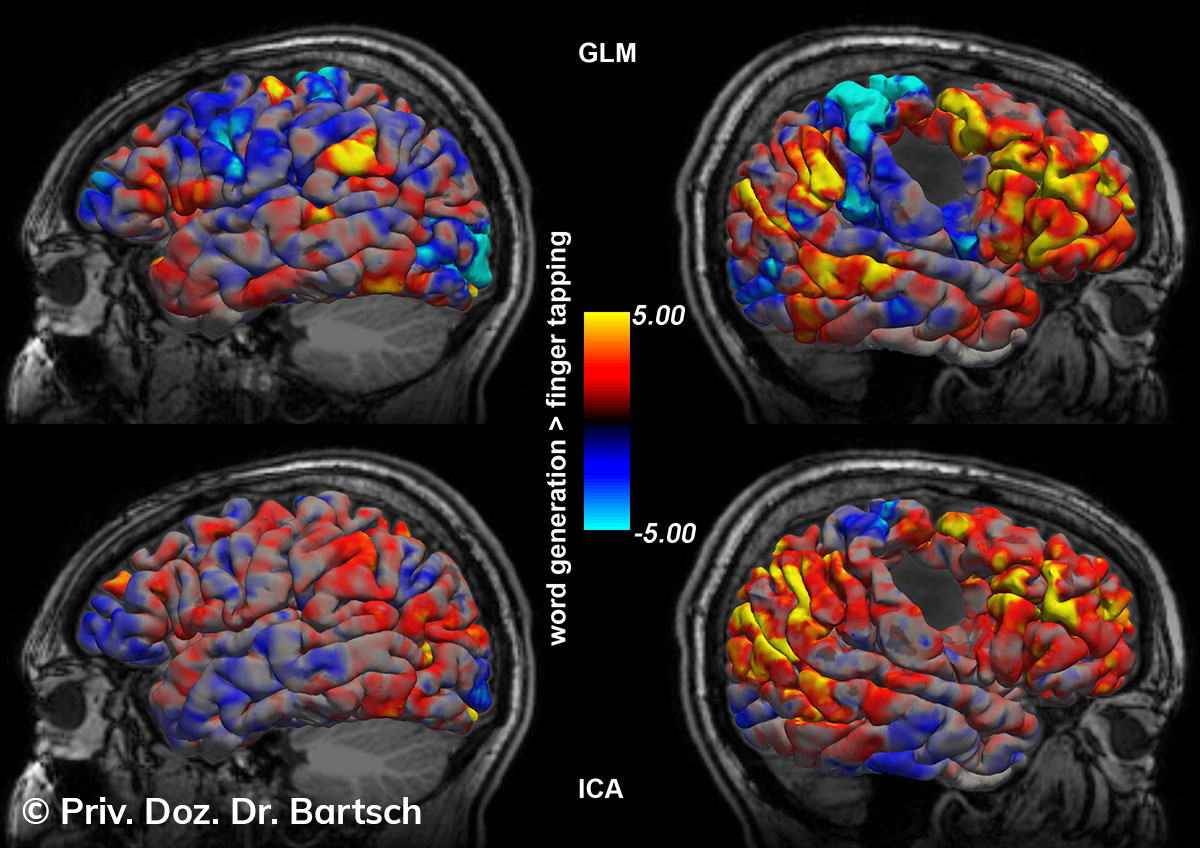
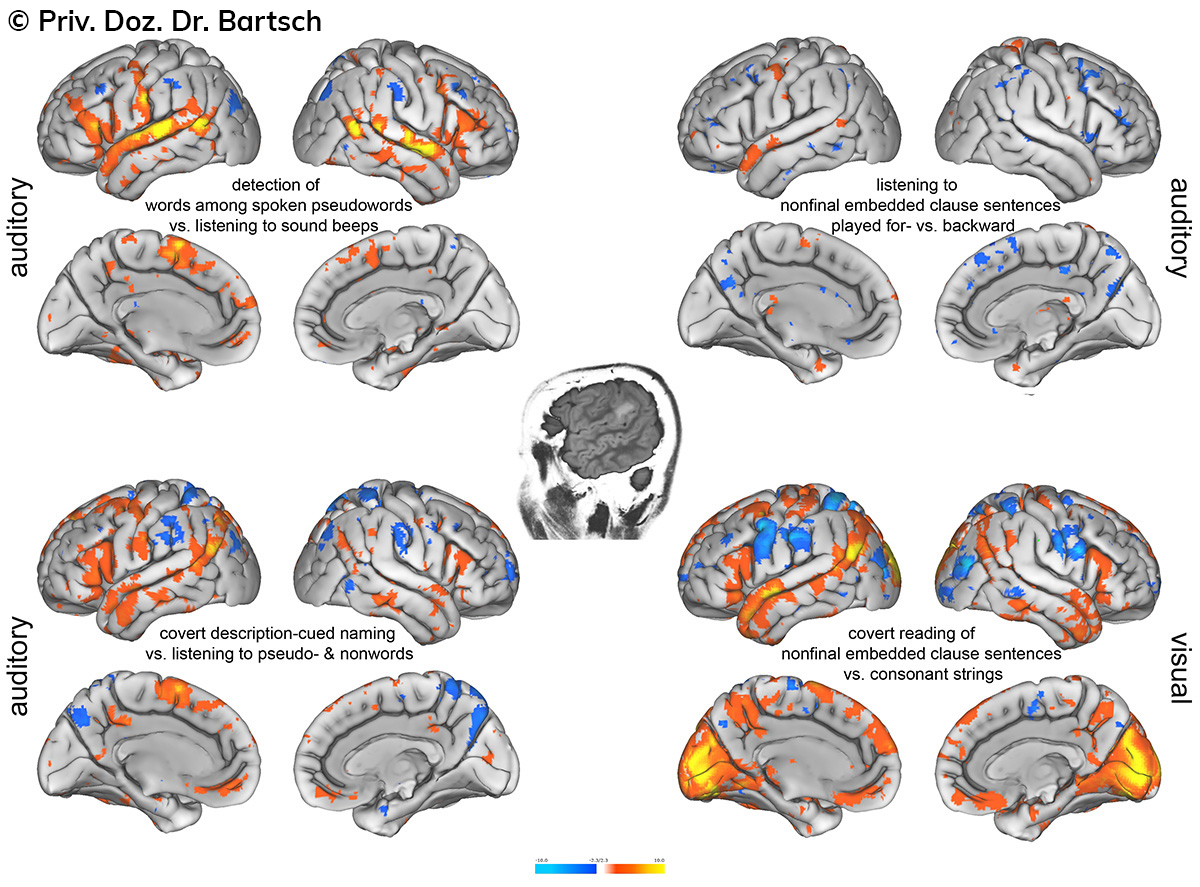
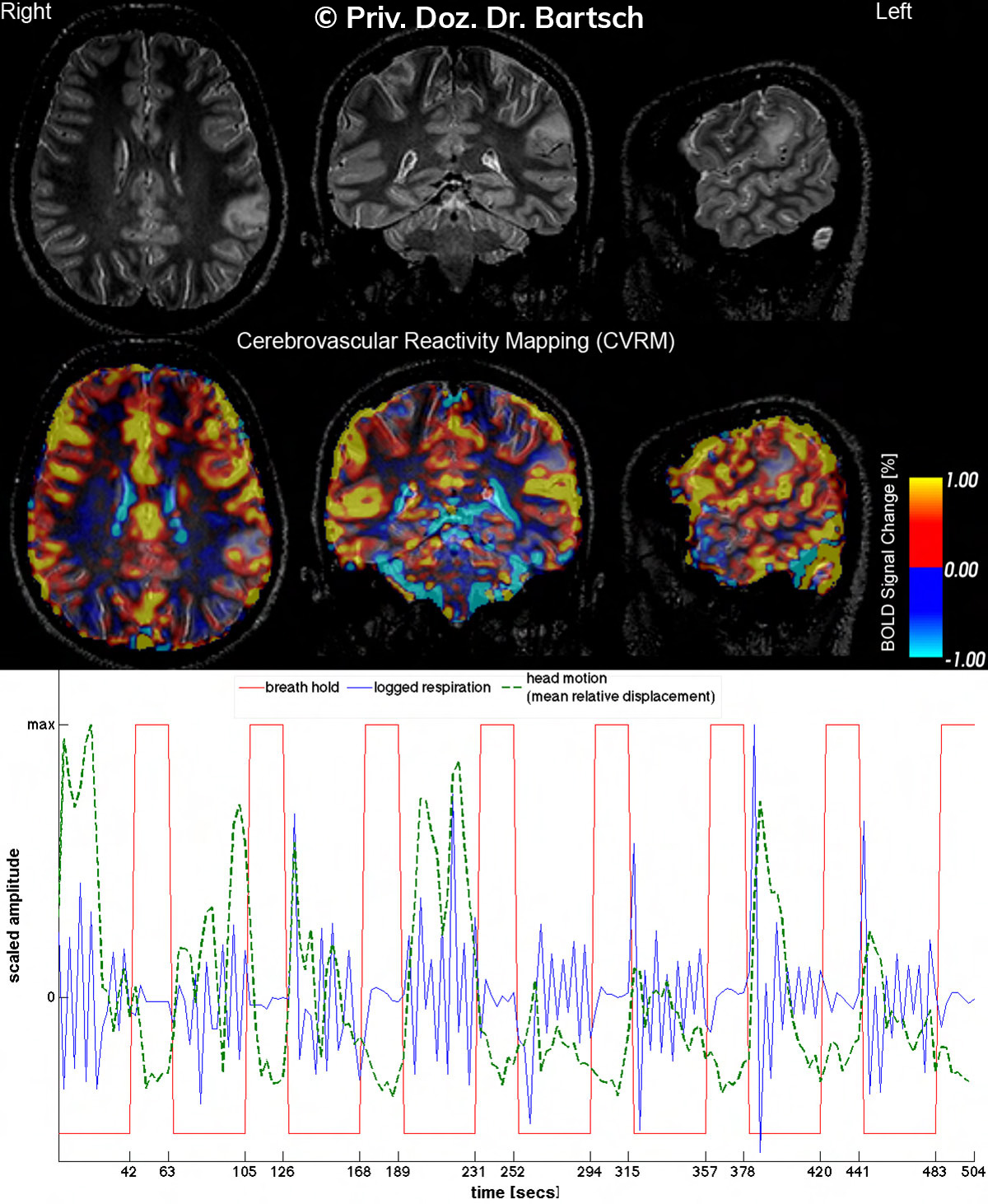
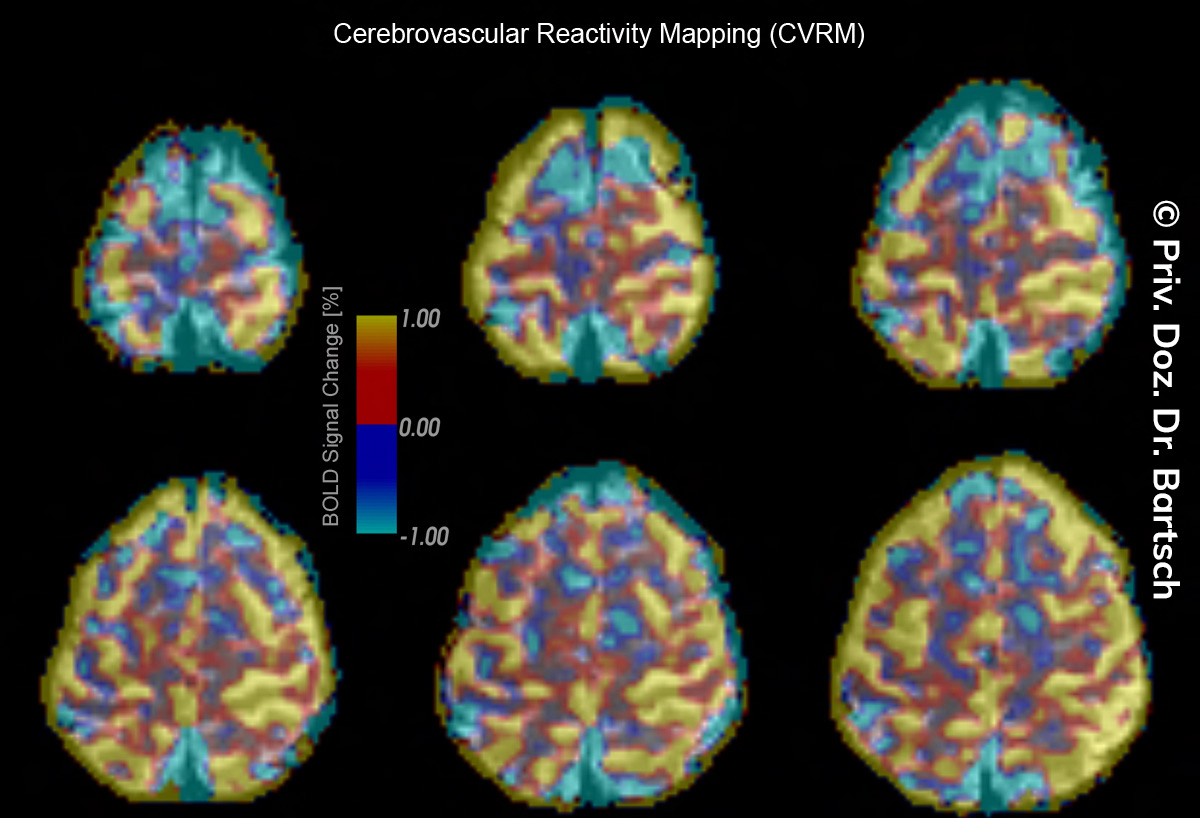
Untersuchungen bei Verdacht auf dementielle Erkrankungen und Multipler Sklerose
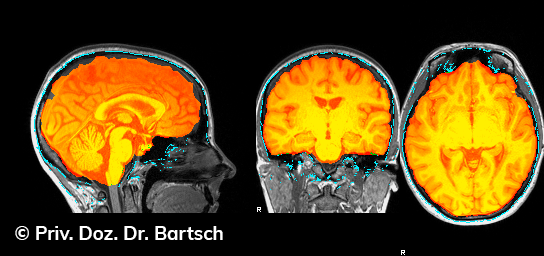
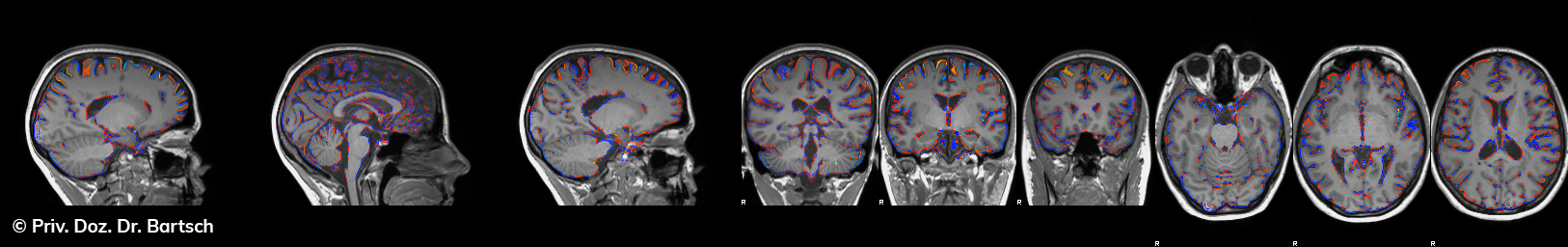
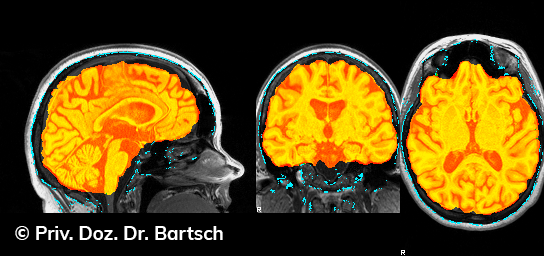
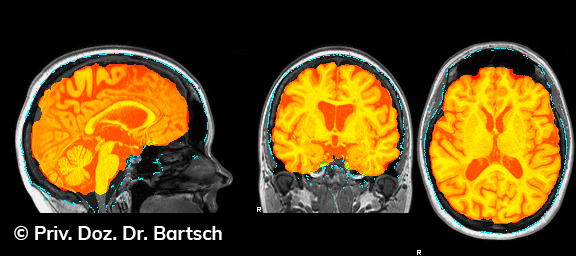
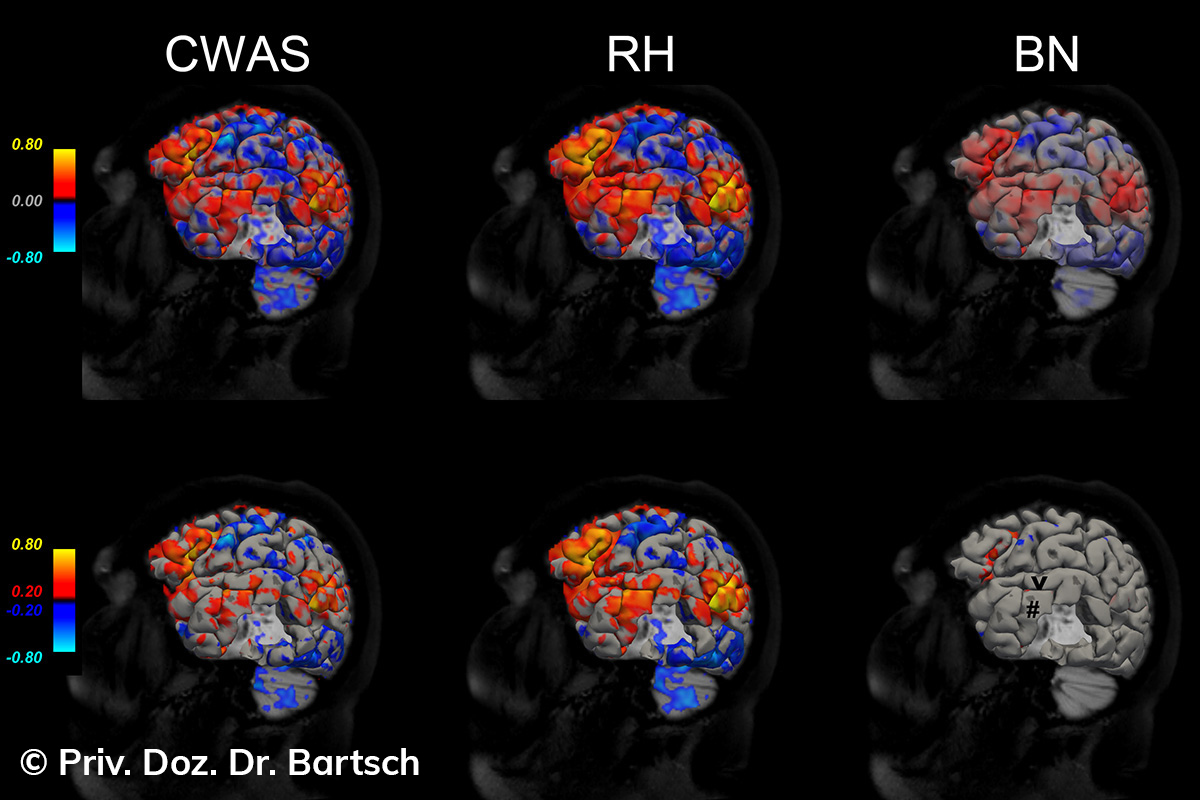
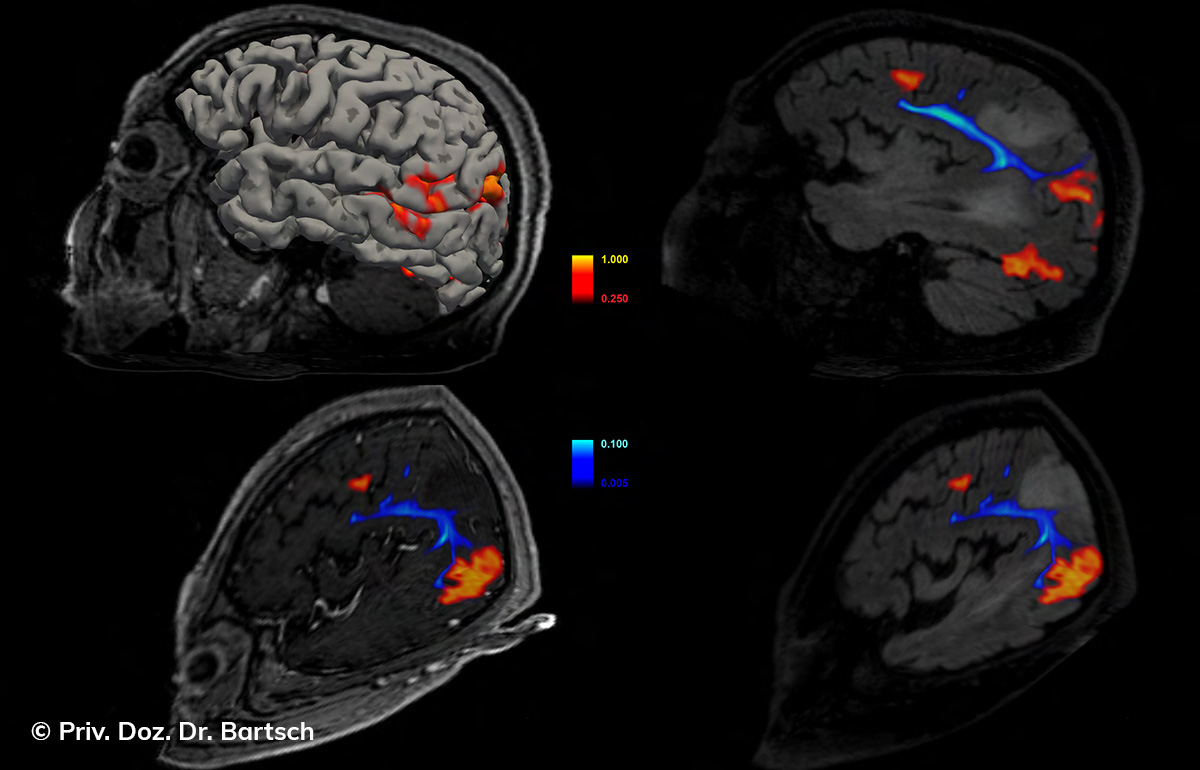
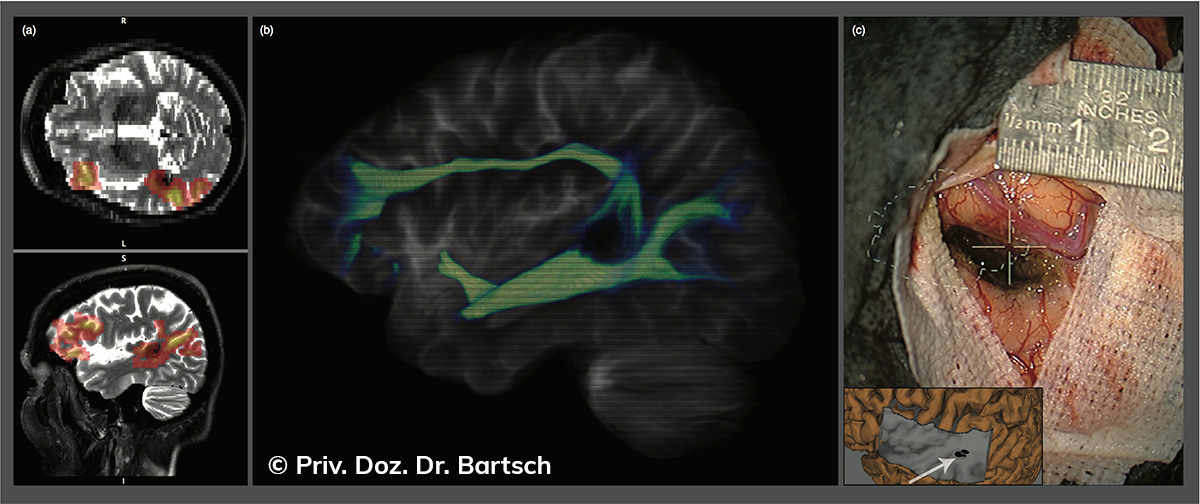
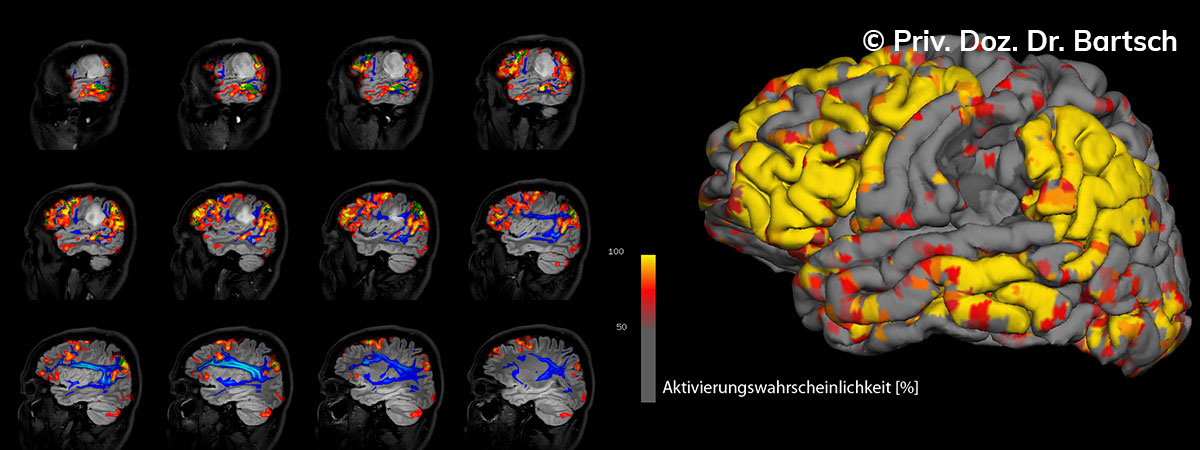
Die Bestimmung des gesamten Hirnvolumens und des Volumens der grauen, Nervenzellkörper beinhaltenden Substanz bzw. ausgewählter Bereiche des Gehirns (die sogenannte zerebrale Morphometrie) gewinnt in der Diagnostik und bei Verlaufsuntersuchungen von Patienten mit neurodegenerativen (wie der Alzheimer-Demenz) und entzündlichen ZNS-Erkrankungen (wie der Multiplen Sklerose) zunehmend an Bedeutung.
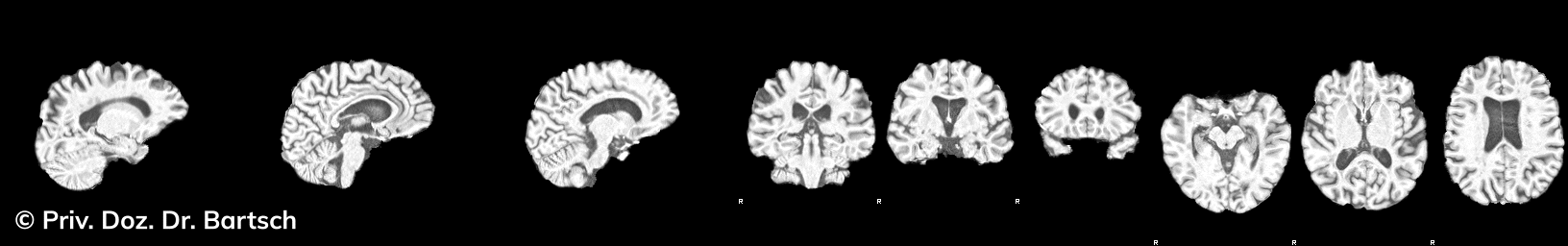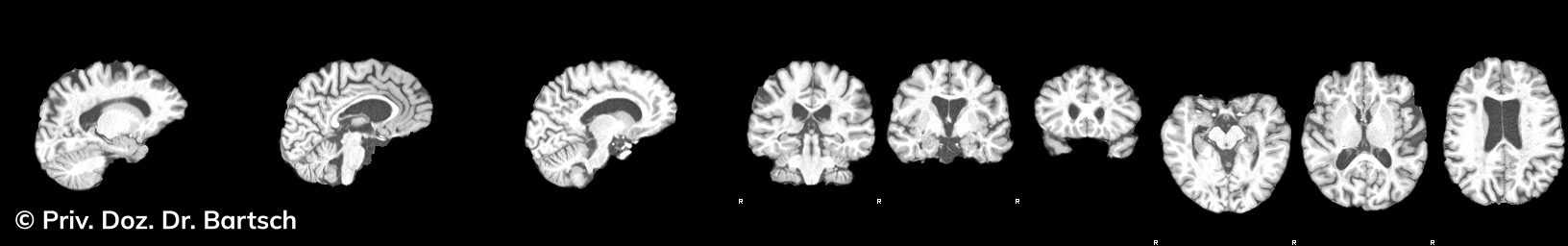
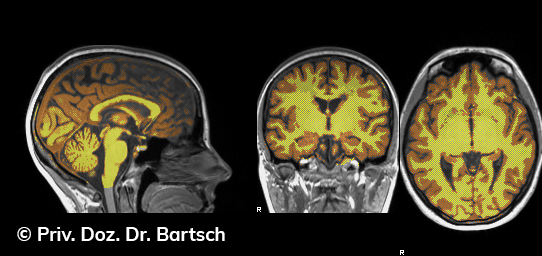
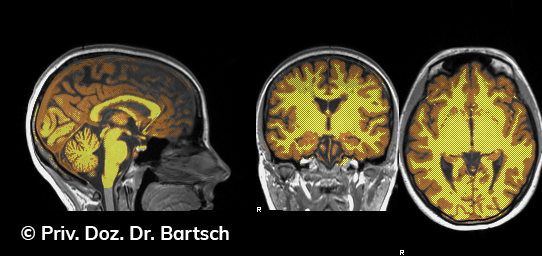
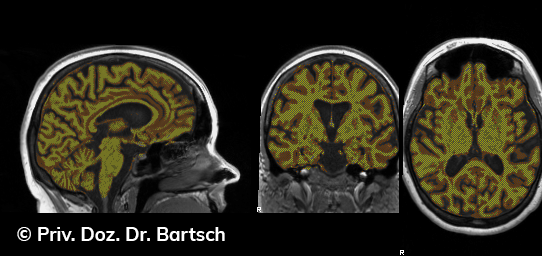
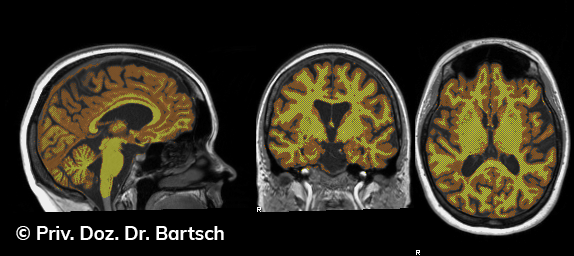
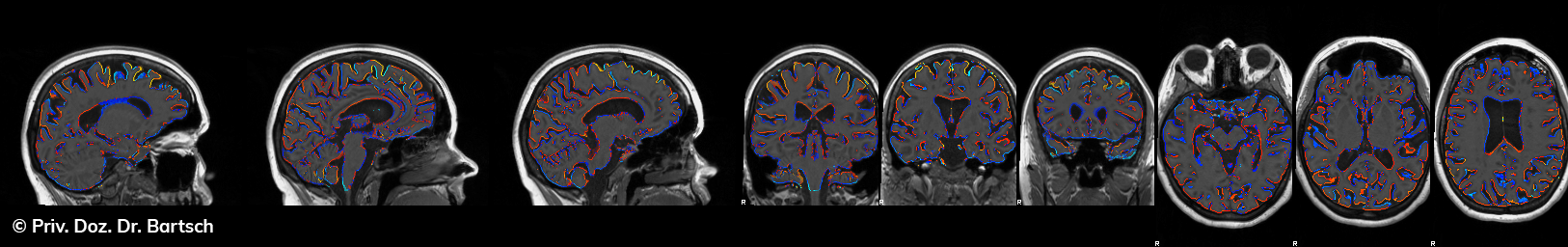
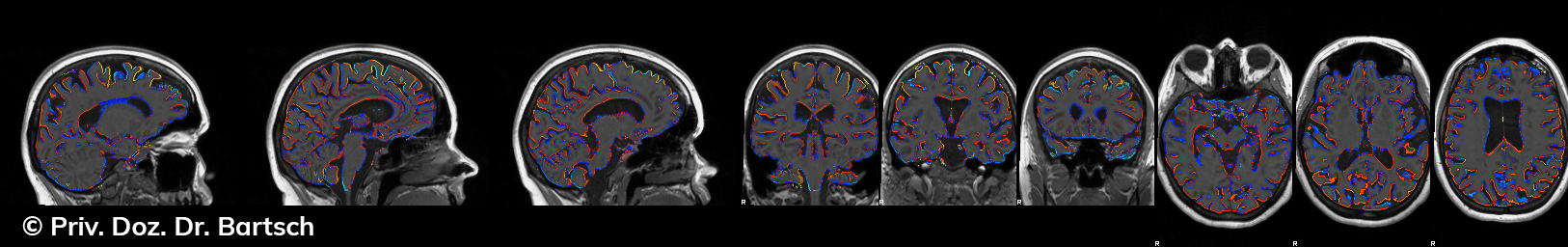
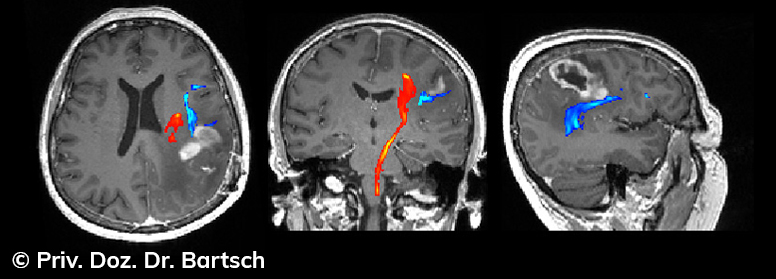
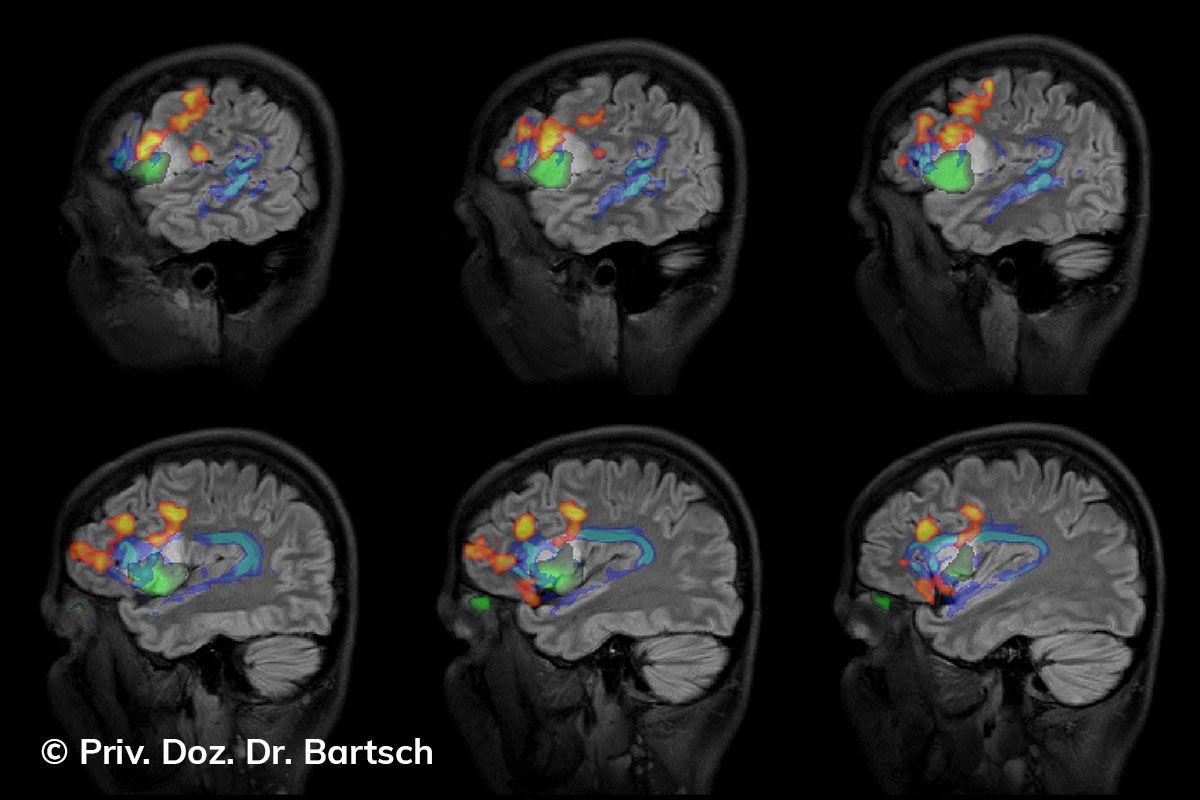
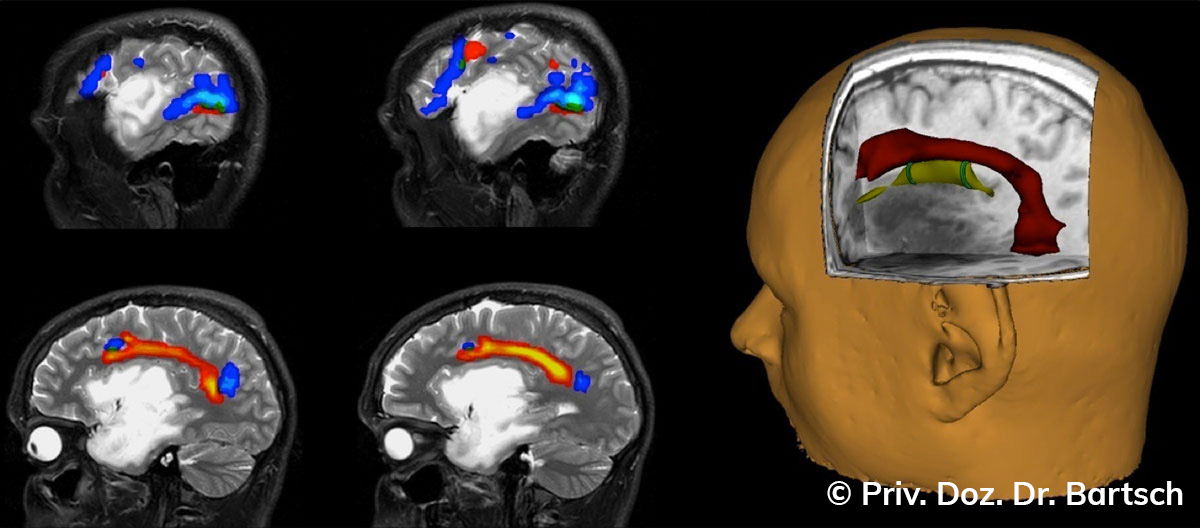
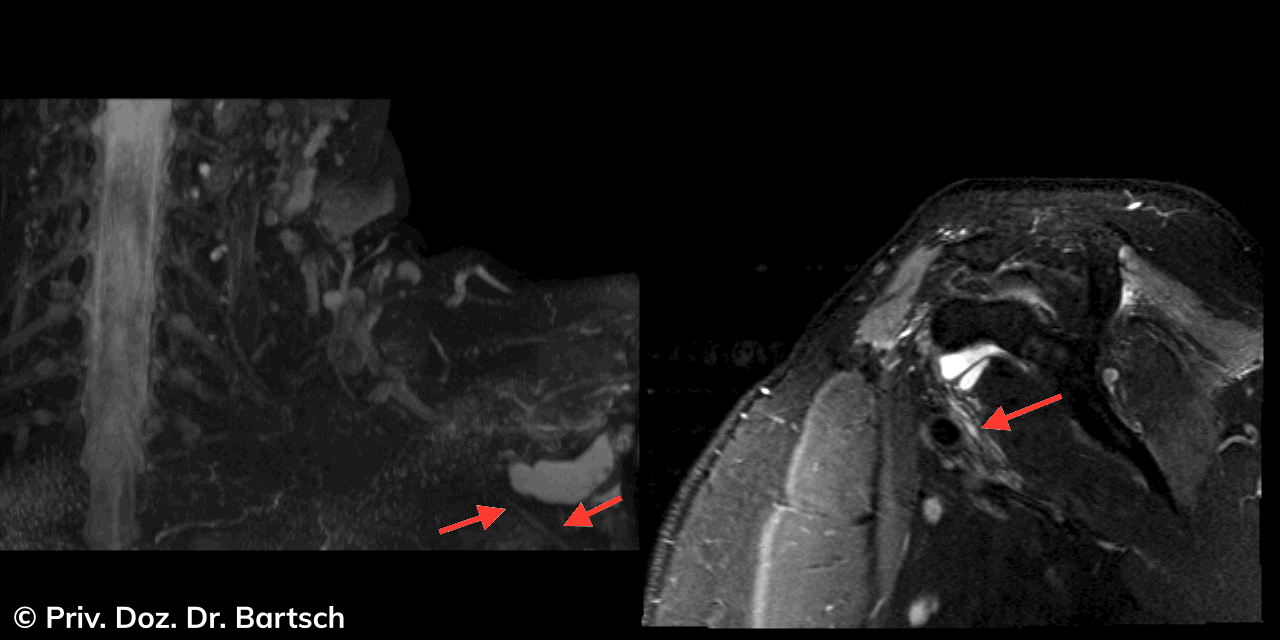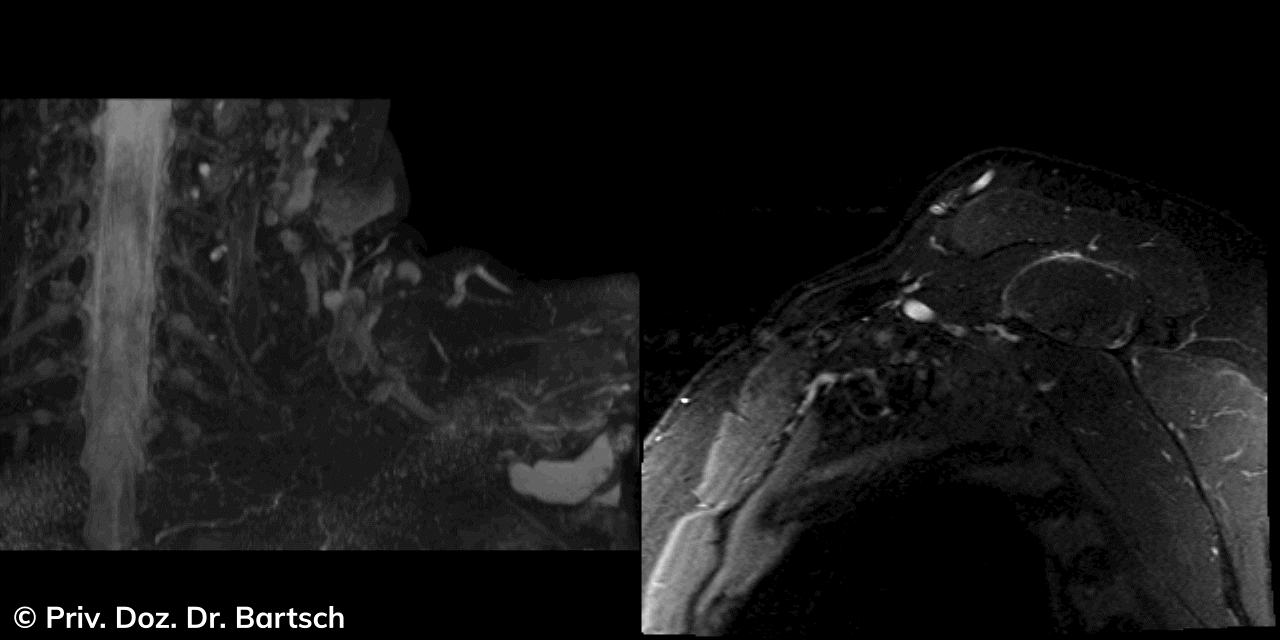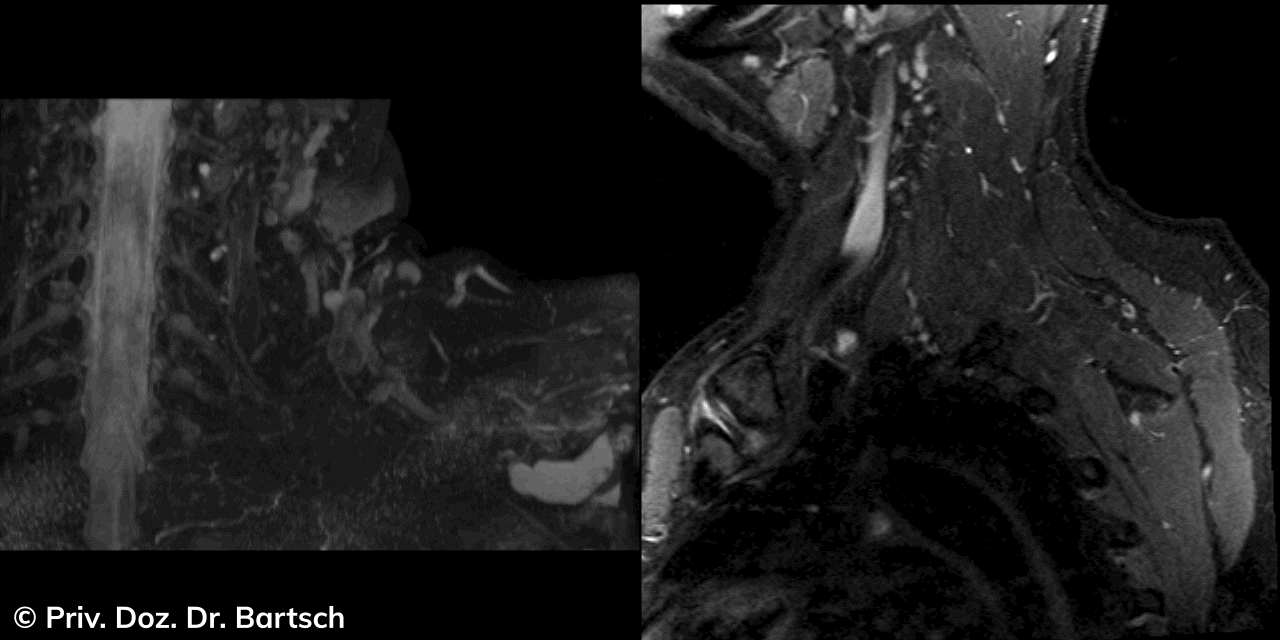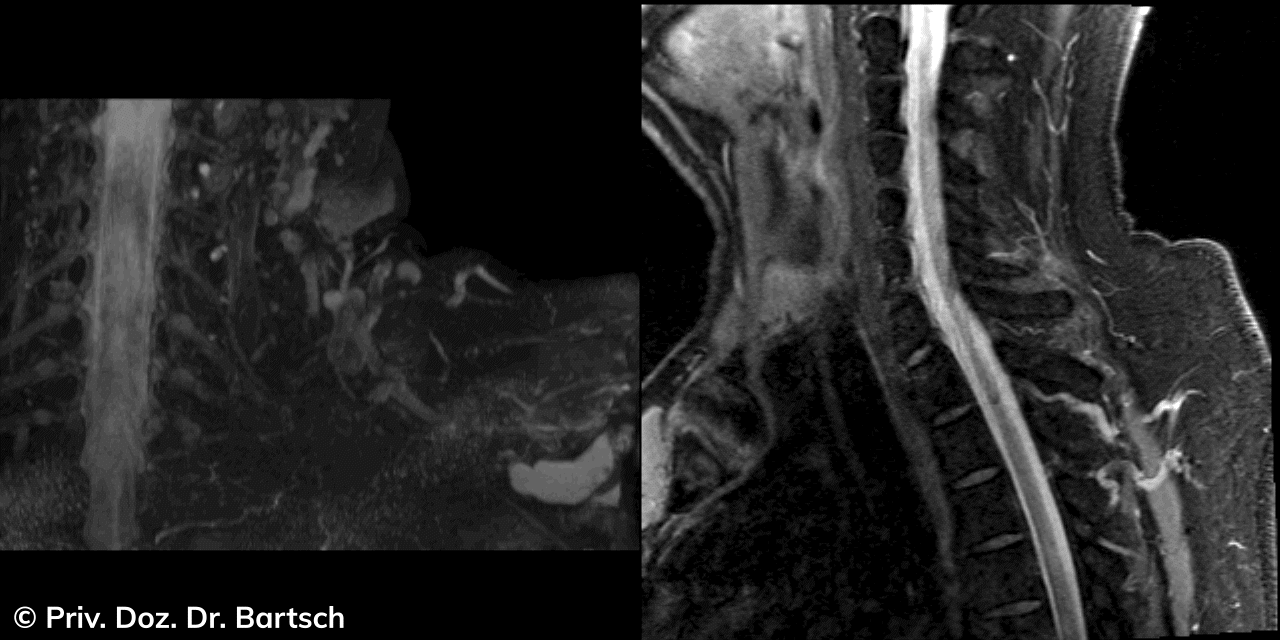
Hauptmerkmal der Multiplen Sklerose sind zeitlich und örtlich an bevorzugten Stellen verstreut auftretende Entmarkungsherde der weißen, Nervenfasern beinhaltenden Hirnsubstanz. Ein Hirnvolumenverlust begleitet jedoch oft die Erkrankung und ist mit dem Auftreten körperlicher und kognitiver Einschränkungen vergesellschaftet. Wenn es dagegen gelingt, einen Hirnvolumenverlust unter der Gabe von Medikamenten zu stoppen, so kann das die Wirksamkeit der Behandlung für den Patienten belegen. Bei beiden hier gezeigten Fällen handelt es sich um etwa 35jährige Frauen, die seit circa 5 Jahren an Multipler Sklerose erkrankt waren.
Im Fall 1 (MS-Beispielfall 1 Morphometrie) kam es im Verlauf eines Jahres unter der Behandlung mit einem neuen Medikament zu keiner wesentlichen Hirnvolumenänderung. Im Fall 2 (MS-Beispielfall 1 Morphometrie) nahm das Hirnvolumen im gleichen Zeitraum dagegen um mehr als 0.52 % ab. Das zeigt in mehr als 95 % der Fälle einen fortschreitenden Abbauprozeß an (J Neurol Neurosurg Psychiatry 2015;0:1–7) und kann dazu anregen, die weitere Behandlung zu überdenken und zu optimieren. Die Radiologie Bamberg bietet derartige Untersuchungen und komplexe Auswertungen für ausgewählte Fragestellungen in enger Kooperation mit den klinisch-fachärztlichen Spezialisten an und ist auch an verschiedenen multizentrischen Studien beteiligt.